Skin Lesion Project
Skin Lesion Detection, Texture Analysis, and Malignancy Classification
Introduction
The goal of the project is to develop and thoroughly evaluate robust, automated texture feature extraction and selection, as well as learning algorithms for classifying pigmented skin lesions to improve the diagnosis of skin cancer in primary care centers. This project aims to improve skin cancer detection and care. A key objective of the project is to develop an interactive support tool with straight forward GUI to assist primary care clinicians in the accurate screening of pigmented skin lesions to improve early detection, reduce unnecessary referrals to hospitals and reduce the number of missed malignant melanomas.
The impact of the proposed project is twofold: (i) significantly improve understanding of correspondence between automated extracted features for computerized automated identification (such as size, shape, color, and texture information) and the ABCD rule of thumb physicians used to pre-screening the early skin cancer; and (ii) advances integration of feature extraction and classification in bio-medical imaging processing.
The overarching goal is to provide transparent support for skin cancer screening by primary care physicians based on advanced bio-medical image processing and decision making techniques.
Application Architecture and Flowchart
The main functional requirements for an early melanoma detection system (1) well designed GUI that provides physicians the flexibility to access detailed methodology or just use the diagnostic information recommended with confidence level; (2) robust and efficient image processing, feature extraction, and classification algorithms designed for skin lesion imaging processing.
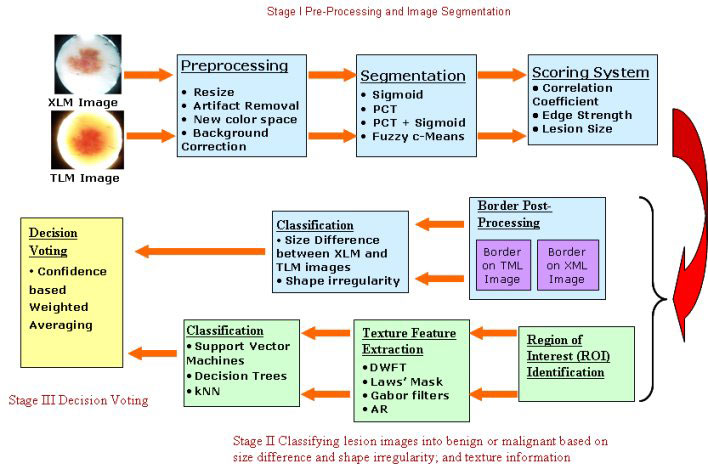
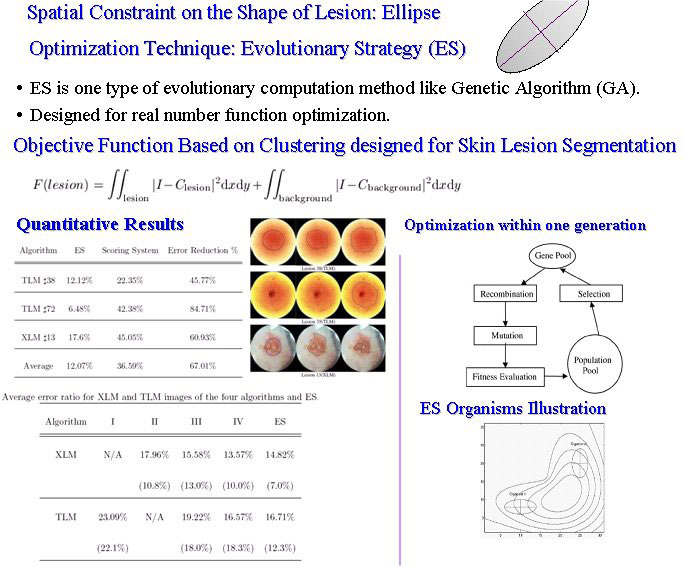
Skin Lesion Detection using ES-based segmentation algorithm
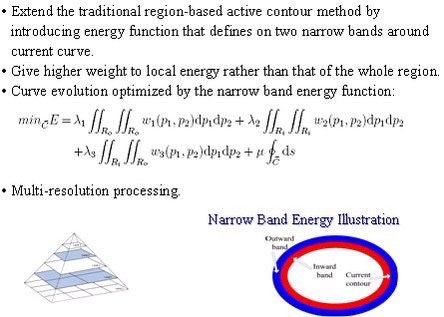
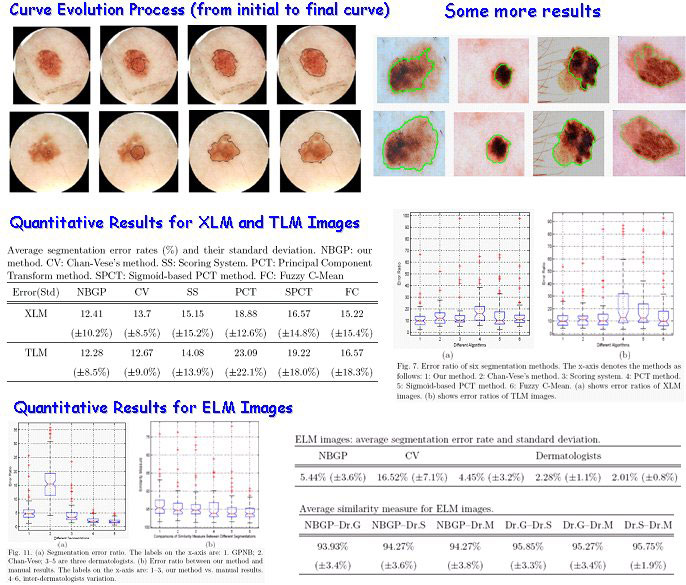
Skin Lesion Detection using narrow band graph partitioning algorithm
Skin Lesion Texture Analysis
GUI Design
Following figures show GUI design for pre-processing and segmentation steps of the system. The system will be used by clinicians and we expect the GUI will change to fit their taste.
Following Figure shows available pre-processing options and segmentation results.
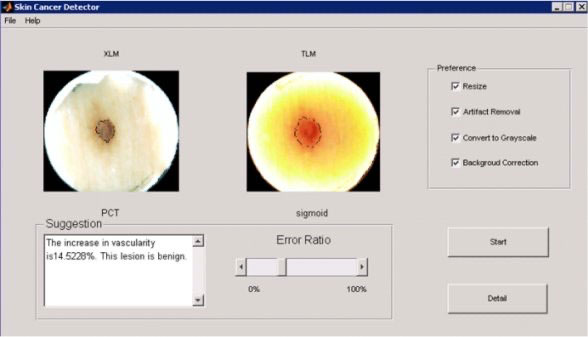
The figure below also provide user flexibility to choose different segmentation algorithms manually.
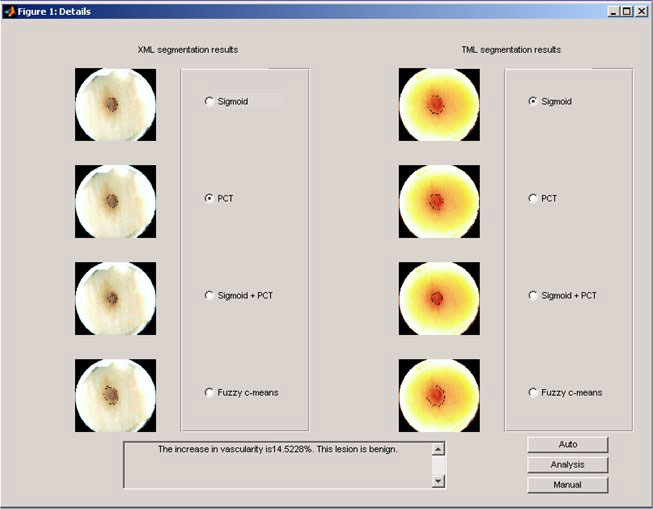
More detailed information, such as result analysis for texture feature extraction and selection, ROC analysis for classifier selection can be found in the published work.
Graduate Students working for the project: Zhenyu Yang (graduated), Ning Situ
Collaborators: Drs. George Zouridakis (ET and TLC2) and Ji Chen (ECE)